Professor, MCB
Degrees and Appointments
- B.S. 1986, Southern Illinois University, Carbondale
- Ph.D. 1994, University of Pennsylvania, Philadelphia
- Postdoc 1994-1996, The Upjohn Company
- Postdoc and Fellow 1996-1999, University of California, Santa BarbaraAssistant Researcher I 1999-2001, University of California, Santa Barbara
Fields of Study: Biochemistry
Research Specialities: Bioanalytical, Biophysics, Chemical Reaction Dynamics/Kinetics/Interactions, Metabolism, Signaling, and Regulation, Nucleic Acids and Genomes, Protein and Membrane Biochemistry, Spectroscopy/Molecular Structure, Structural Biology, Theory, Modeling, and Simulation
Awards and Honors:
- JSPS International Fellowship for Research in Japan, 2019
- Contributing member in DNA Replication and Repair, Faculty of 1000, 2010-pres
- Nielsen Trust Fund Award, College of Medicine, 2001
- NSF POWRE Award, 1999-2001
- American Cancer Society Postdoctoral Fellow, 1997-1999
Research
Phage-host systems are under intense evolutionary pressure, consequently they have developed remarkably ingenious mechanisms of attack and defense. Our lab investigates one such remarkable system: that found in Streptomyces griseus. Based on its biochemical activities, SgrAI, a nucelase from S. griseus, is postulated to be activated by binding to invading phage DNA, simultaneously expanding its DNA sequence cleavage specificity and forming polymers that may act to protect the host DNA from its resulting off-target cleavage activity. Enzyme mechanisms involving polymer or filament formation are exceedingly rare, although recent screens suggest this may be more common than previously thought. Being a potentially new paradigm for enzyme regulation, several fundamental questions arise that are under investigation in our lab, including the structure, kinetics, and biological role of the polymer. Our previous biochemical data suggests that the polymer formed from activated SgrAI is a run-on oligomer, which has now been confirmed by the 8.6 Å cryo-electron microscopy structure determined in our lab. Although this structure shows how the SgrAI dimers bound to activating DNA associate in a repeating helical arrangement, fundamental questions such as how DNA cleavage is activated, how DNA sequence specificity is altered, and whether or not domain swapping (found in a crystal structure of two DNA bound SgrAI dimers) is present require higher resolution and therefore remain to be answered. Also important to understanding the function of the run-on oligomer is determining how formation of such an assembly, where the bound DNA appears critical for oligomer stability, accelerates rather than impedes multiple DNA cleavages. Finally, the biological role for run-on oligomer formation has been hypothesized to function in protecting the host DNA from dangerous off-target cleavages made possible via activation of SgrAI, by sequestering SgrAI on the invading phage DNA. Current studies in our lab aim to investigate the structure of the run-on oligomer using biochemical and x-ray crystallographic methods, measure kinetic steps involving polymer formation and dissociation in the reaction pathway using pre-steady state fluorescence methods, and test the postulated biological role of the polymer using in vitro and in vivo assays including phage infection challenges.
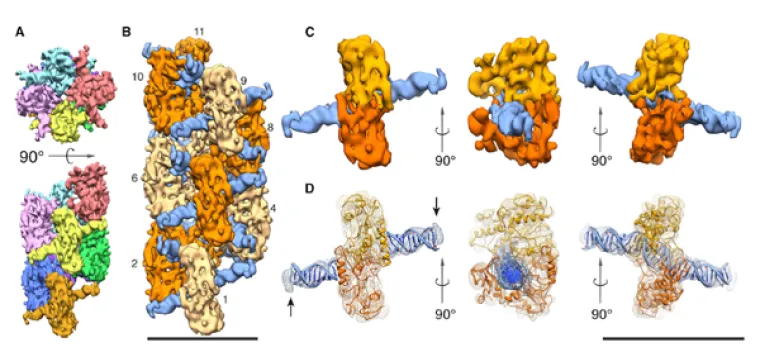
Single particle reconstruction of SgrAI/DNA polymer using cryo-electron microscopy (CryoEM). A. Top and side views showing the helical organization of the SgrAI polymer. Eight distinct DNA binding domains (DBD) are shown in distinct colors. B. Helical reconstruction of the SgrAI/DNA polymer at 8.6 Å resolution using cryoEM data. SgrAI shown as light and dark orange, DNA shown as dark blue. C. CryoEM envelope around one DNA bound SgrAI dimer, shown from three different views. D. As in C with transparency and the ribbon drawing of the fitted coordinates of SgrAI bound to DNA derived from x-ray crystallography. The two subunits of the SgrAI dimer from the polymer undergo a 10° rotation relative to the x-ray crystallographic structure of the free, unpolymerized SgrAI/DNA complex. (Adapted from Lyumkis, et al., 2013, Structure 21, 1848-1858.)
Triggering of Autoimmune Disease by Viral Infection
Human Parvovirus B19 (B19V) is a small single stranded DNA virus that infects the majority of the population. Though most individuals show no lasting effects from the viral infection, several very serious conditions (aplastic crisis, pure red-cell aplasia, and hydrops fetalis) are known to occur in susceptible populations, and many instances of liver, heart, and autoimmune disease following infection have also been reported. Yet, despite the ubiquity of the virus and the rare but serious complications, few detailed molecular studies have been performed with B19V or its components. We propose to begin such studies by investigating the roles of the main B19V replication protein, NS1, and its role in two biological processes, namely viral replication and host gene transactivation. Of interest is how NS1 accomplishes these processes which include specific recognition of DNA and host proteins, as well as manipulation of DNA including cleavage and alterations of DNA structure. These studies will make use of both purified, recombinant NS1 protein, as well as NS1 expressed in different human cell lines, and will build the groundwork for future studies in elucidating the role of NS1 in the different disease states, as well as how interactions with different cellular environments, including different genetic backgrounds, leads to these disease states.
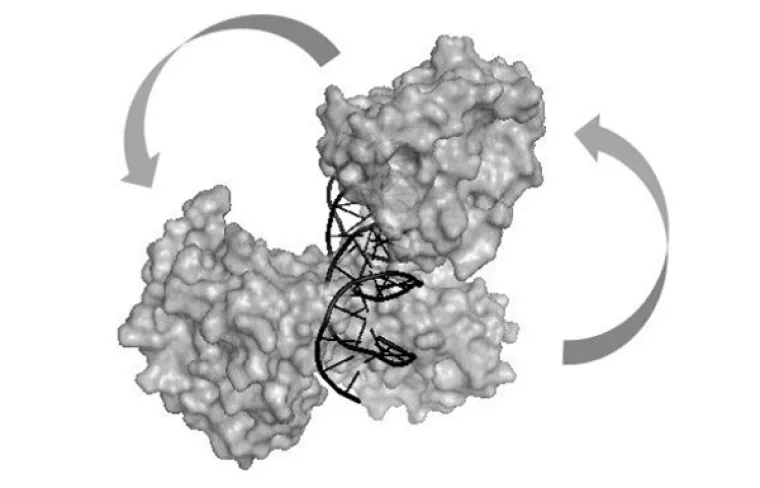
Model of human parvovirus B19 NS1 protein nuclease domains interacting with duplex DNA.
- Zerio, C.J., Sivinski, J., Wijeratne, E.M.K., Xu, Y.M., Ngo, D.T., Ambrose, A.J., Villa-Celis, L., Ghadirian, N., Clarkson, M.W., Zhang, D.D., Horton, N.C, Gunatilaka, A.A.L., Fromme, R., Chapman. E.J. (2023) “Physachenolide C is a Potent, Selective BET Inhibitor” J. Med. Chem. 2023, 66, 1, 913–933. https://doi.org/10.1021/acs.jmedchem.2c01770. doi: 10.1021/acs.jmedchem.2c01770. PMID: 36577036.
- Lyumkis, D & Horton, N.C. (2022) "The role of filamentation in activation and DNA sequence specificity of the sequence-specific endonuclease SgrAI", (2022)Biochem Soc Trans 50 (6): 1703–1714. https://doi.org/10.1042/BST20220547.
Sanchez, J.L., Ghadirian, N., & Horton, N.C. (2022) “Structure of the N-terminal Nuclease and Origin Binding Domain of Human Parvovirus B19 Main Replicative Protein NS1”, J. Virology, PMID: 35435730, DOI: 10.1128/jvi.02164-21. - Shan, Z., Ghadirian, N., Lyumkis, D. and Horton, N. C. (2022) "Pre-Transition State and Apo Structures of the Filament-Forming Enzyme SgrAI Elucidate Mechanisms of Activation and Substrate Specificity", J. Biol. Chem., 298, 10176. (PMID: 35202658. DOI: 10.1016/j.jbc.2022.101760).
- Townsend, J., Sanders, H., Rolland, A., Park, C., Horton, N., Prell, J., Wang, J., Marty, M. (2021) "Influenza A M2 Channel Oligomerization is Sensitive to its Chemical Environment", Analytical Chem. 93, 16273-16281, doi: 10.1021/acs.analchem.1c04660, PMID: 34813702.
- Park, C. K. & Horton, N.C. (2020) “Novel insights into filament forming enzymes”, Nature Reviews Molecular Cell Biology, 21(1):1-2. doi: 10.1038/s41580-019-0188-1.
- Horton, N.C. (2020) “The Filament Forming Mechanism of SgrAI Endonuclease‐Structural and Kinetic Analysis”, The FASEB Journal 34 (S1), 1-1. https://doi.org/10.1096/fasebj.2020.34.s1.04300
- Horton, N.C. (2020) “Filament Formation Induces a Shape Change and Activation of the Nuclease SgrAI”, The FASEB Journal 34 (S1), 1-1. https://doi.org/10.1096/fasebj.2020.34.s1.00728
- Ghadirian, N., Horton, N. (2020) “Structure‐Function Studies of the Helicase Domain of NS1 Protein of Human Parvovirus B19”, The FASEB Journal 34 (S1), 1-1. https://doi.org/10.1096/fasebj.2020.34.s1.00730
- Park, C. K. & Horton, N.C. (2019) “Structures, Functions, and Mechanisms of Filament Forming Enzymes: A Renaissance of Enzyme Filamentation”, Biophysical Reviews, 11(6):927-994. doi: 10.1007/s12551-019-00602-6.
- Polley, S., Lyumkis, D., and Horton, N. C. (2019) “Mechanism of Filamentation-Induced Allosteric Activation of the SgrAI Endonuclease”, Structure 27, 1-11.
- Barahona, C., Basantes, L. E., Tomkins, K. J., Heitman, D. M., Chukwu, B. I., Sanchez, J., Sanchez, J. L., Ghadirian, N., Park, C. K., and Horton, N. C. (2019) “The Need for Speed: Run-On Oligomer Filament formation provides Maximum Speed with Maximum Sequestration of Activity”, J. Virol, 93 (5), e01647-18. doi: 10.1128/JVI.01647-18. PMID: 30518649.
- *Featured on the cover
- Xu, P., Ganaie, S., Wang, X., Wang, Z., Kleiboeker, S., Horton, N.C., Heier, R., Meyers, M., Tavis, J., and Qiu, J. (2019) "Endonuclease Activity Inhibition of the NS1 Protein of Parvovirus B19 as a Novel Target for Antiviral Drug Development", Antimicrob Agents Chemother, 63 (3), e01879-18. doi: 10.1128/AAC.01879-18. PMID: 30530599.
- Park, C. K. & Horton, N.C. (2019) “Structures, Functions, and Mechanisms of Filament Forming Enzymes: A Renaissance of Enzyme Filamentation”, arXiv: arXiv:1909.13141 [q-bio.BM].
- Polley, S., Lyumkis, D., and Horton, N. C. (2019) “Indirect Readout of DNA Controls Filamentation and Activation of a Sequence-Specific Endonuclease”, bioRxiv, DOI: https://doi.org/10.1101/585943.
- Horton, N.C., Park, C.K., Barahona, C., Basantes, L.E., & Ghadirian, N. (2019) “Kinetic Advantages of the Run-On Oligomer or Filamentation Mechanism of a DNA Cleaving Enzyme”, FASEB J., 33 (1_supplement), 633.12-633.12.
- Park, C. K., Sanchez, J. L., Barahona, C. J., Basantes, L. E., Sanchez, J., Hernandez, C., and Horton, N. C. (2018) “The Run-on Oligomer Filament Enzyme Mechanism of SgrAI. Part 1: Assembly Kinetics of the Run-on Oligomer Filament”, J. Biol. Chem., 293(38):14585-14598.
- Park, C. K., Sanchez, J. L., Barahona, C. J., Basantes, L. E., Sanchez, J., Hernandez, C., and Horton, N. C. (2018) “The Run-on Oligomer Filament Enzyme Mechanism of SgrAI. Part 2: Kinetic Modeling of the Full DNA Cleavage Pathway”, J. Biol. Chem., 293(38):14599-14615.
- Horton, N., Park, C., Sanchez, J., Barahona, C., & Basantes, L.E. (2018) “Enzyme activity and specificity modulated by protein filamentation”, Prot. Sci., 27, 117-11.
- Hernandez, C.J., Sanchez, J.L. & Horton, N.C. (2017) “Investigating the role of the human parvovirus B19’s main viral protein, NS1, in viral replication and interactions with host DNA,” (2017) FASEB J. 31, 912.16-912.16
- Sanchez , J.L., Romero, Z., Quinones, A., Torgeson, K.R. & Horton, N.C. (2016) “DNA Binding and Cleavage by Human Parvovirus B19 NS1 Nuclease Domain”, Biochemistry, 55, 6577-6593. PMID: 27809499.
- Shah, S., Sanchez, J., Stewart, A., Piperakis, M.A., Cosstick, R., Nichols, C., Park, C.P., Ma, X., Wysocki, V., Bitinaite, J. & Horton, N.C. (2015) “Probing the Run-on Oligomer of Activated SgrAI bound to DNA”, PLoS One, Apr 16;10(4):e0124783.
- Shah, S, Dunten, P., Stiteler, A., Park, C.K. & Horton, N.C. (2015) “Structure and Specificity of FEN-1 from Methanopyrus kandleri”, Proteins, 83, 188-194.
- Lyumkis, D., Talley, H., Stewart, A., Shah, S., Park, C.K., Tama, F., Potter, C.S., Carragher, B., Horton, N.C. (2013) “Allosteric Regulation of DNA Cleavage and Sequence-Specificity through Run-On Oligomerization”, Structure, 21, 1848-1858.
- Ma, X., Shah, S., Zhou, M., Park, C.K., Wysocki, V.H., Horton, N.C. (2013) “Structural Analysis of Activated SgrAI-DNA Oligomers Using Ion Mobility Mass Spectrometry”, Biochemistry, 52, 4373-81.
- Horton, N.C., Park, C.K., Stewart, A.M., Shah, S., Talley, H., Ma, X., Wysocki, V., Piperakis, M., Cosstick, R. & Jacovetty, E. (2012) “Activation by oligomerization of an allosteric sequence specific endonuclease,” FASEB J. 26, ib91-ib91.
- Little, E.J., Dunten, P.W., Bitinaite, J. & Horton, N.C. (2011) “New Clues in the Allosteric Activation of DNA Cleavage by SgrAI; Structures of SgrAI Bound to Cleaved Primary Site DNA and Uncleaved Secondary Site DNA”, Acta Cryst., 67, 67-74.
- Techner, J.-M. & Horton, N.C. (2011) “Small molecule modulation of zinc-finger sequence specificity,” FASEB J. 25, 688.6-688.6.
- Park, C.K., Stiteler, A.P., Shah, S., Ghare, M.I., Bitinaite, J. & Horton, N.C. (2010) “Activation of DNA Cleavage by Oligomerization of DNA bound SgrAI”, Biochemistry, 49, 8818-8830.
- Park, C.K., Joshi, H.K., Agrawal, A., Ghare, M.I., Little, E.J., Dunten, P.W., Bitinaite, J. & Horton, N.C. (2010) “Domain Swapping in Allosteric Modulation of DNA Specificity”, PLoS Biology, 8(12):e1000554.
- Horton, N.C. & Park, C.K., (2010) “Crystallization of Zinc Finger Proteins bound to DNA”, Methods Mol Biol. 649:457-77.
- Dunten, P.W., Little, E.J. & Horton, N.C. (2009), “The restriction enzyme SgrAI: structure solution via combination of poor MIRAS and MR phases.”, Acta Cryst. D65, 393-8.
- Little, E.J., Babic, A.C., & Horton, N.C. (2008) “Early interrogation and recognition of DNA sequence by indirect readout”, Structure 16, 1828-37. (Given F1000 “must read”)
- Babic, A.C., Little, E.J., Manohar, V.M., Bitinaite, J., & Horton, N.C. (2008) “DNA distortion and specificity in a sequence-specific endonuclease” J. Mol. Biol. 383, 186-204.
- Dunten, P.W., Little, E.J., Gregory, M.T., Manohar, V.M., Dalton, M., Hough, D., Bitinaite, J., Horton, N.C. (2008) “The structure of SgrAI bound to DNA; recognition of an 8 base pair target”, Nucleic Acids Res. 36, 5405-16.
- Horton, N.C. (2008) “Deoxyribonucleases”. In Protein-Nucleic Acid Interactions: Structural Biology; Carl C. Correll, Pheobe Rice, Eds.; RSC Publishing: Cambridge, United Kingdom, 2008.
- Segal, D.J., Crotty, J., Bhakta, M., Barbas III, C.F. & Horton, N.C. (2006) "Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA", J. Mol. Biol. 363, 405-421.
- Joshi, H.K., Etzkorn, C., Chatwell, L., Bitinaite, J., & Horton, N.C. (2006) "Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism." J. Biol. Chem. 281, 23852-69.
- Little, E.J. & Horton, N.C. (2005) "DNA induced conformational changes in type II endonucleases; the structure of unliganded HincII" J. Mol. Biol. 351, 76-88.
- *Featured on the cover.
- Crotty, J.W., Etzkorn, C., Barbas, III, C.F., Segal, D.J. & Horton, N.C. (2005) "Crystallization and preliminary X-ray crystallographic analysis of Aart, a designed six-finger zinc-finger peptide, bound to DNA" Acta. Cryst. F61, 573-576.
- Etzkorn, C. & Horton, N.C. (2004) "Ca2+ Binding in the active site of HincII: Implications for the catalytic mechanism", Biochemistry 43, 13256-70.
- Etzkorn, C. & Horton, N.C. (2004) Mechanistic insights from the structure of HincII bound to cognate DNA cleaved from addition of Mg2+ and Mn2+. J. Mol. Biol. 343, 833-49.
- Horton, N.C., & Perona, J.J. (2004) "DNA cleavage by EcoRV endonuclease: two metal ions in three metal ion binding sites", Biochemistry 43, 6841-57.
- Horton, N.C., Otey, C., Lusetti, S., Sam, M. D., Kohn, J., Martin, A. M., Ananthnarayan, V., & Perona, J.J. (2002) "Electrostatic Contributions to Site Specific DNA Cleavage by EcoRV Endonuclease", Biochemistry 41, 10754-10763.
- Horton, N.C., Dorner, L.F., & Perona, J.J. (2002) "Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation", Nature Struct. Biology 9, 42-47.
- Horton, N.C., & Perona, J.J. (2001) "Making the most of metal ions", Nature Struct. Biology, 8, 290-293.
- Sam, M.D., Horton, N.C., Nissan, T.A. & Perona, J.J. (2001) "Catalytic efficiency and sequence selectivity of a restriction endonuclease modulated by a distal manganese ion binding site", J. Mol. Biol. 306, 851-861.
- Horton, N.C., Connolly, B.A. & Perona, J.J. (2000) "Mechanism of inhibition of phosphoryl transfer in EcoRV by 3’S phosphorothiolates", JACS 122, 3314-3324.
- Horton, N.C. & Perona, J.J. (2000) "Crystallographic snapshots along a protein-induced DNA bending pathway", Proc. Natl. Acad. Sci, USA 97, 5729-5734.
- Perona, J.J., Horton, N.C., Connolly, B.A. & Sam, M.D. (2000) “Catalytic mechanism of EcoRV endonuclease derived from crystal structures and transient kinetics,” Trans. Amer. Cryst. Assoc. 35, 9-
- Horton, N.C., Dorner, L.F., Schildkraut, I. & Perona, J.J. (1999) “Crystallization and preliminary diffraction analysis of the HincII restriction endonuclease –DNA complex”, Acta Cryst. D. 55, 1-3.
- Martin, A.M., Horton, N.C., Lusetti, S., Reich, N.O., & Perona, J.J. (1999) “Divalent metal dependence of site-specific DNA binding in EcoRV endonuclease”, Biochemistry 38, 8430-8439.
- Horton, N.C., Newberry, K.J. & Perona, J.J. (1998) “Metal ion mediated substrate-assisted catalysis in type II restriction endonucleases”, Proc. Natl. Acad. Sci., USA 95, 13489-13494.
- Horton, N.C. & Perona, J.J. (1998) “Recognition of flanking DNA sequences by EcoRV endonuclease involves alternative patterns of water-mediated contacts”, J. Biol. Chem. 273, 21721-21729.
- Horton, N.C. & Perona, J.J. (1998) “Role of protein-induced bending in the specificity of DNA recognition: Crystal structure of EcoRV endonuclease complexed with d(AAAGAT)+d(ATCTT)”, J. Mol. Biol. 277, 779-787.
- Baldwin, E.T., Sarver, R.W., Bryant, G.L.,Jr., Curry, K.A., Fairbanks, M.B., Finzel, B.C., Garlick, R.L., Heinrikson, R.L., Horton, N.C., Kelley, L.C., Mildner, A.M., Moon, J.B., Mott, J.E., Mutchler, V.T., Tomich, C.C., Watenpaugh, K.D., & Wiley, V.H. (1998) “Cation binding to the integrin CD11b I domain and activation model assessment”, Structure 6, 923-935.
- Horton, N.C., Lewis, M. & Lu, P. (1997) “Escherichia coli lac repressor-lac operator interaction and the influence of allosteric effectors”, J Mol. Biol. 265, 1-7.
- Horton, N.C. & Finzel, B.C. (1996) “The structure of an RNA/DNA hybrid: a substrate of the ribonuclease activity of HIV-1 reverse transcriptase”, J. Mol. Biol. 264, 521-533.
- Kercher, M.A., Chang, G., Horton, N.C., Lu, P., Miller, J.H., Pace, H.C. & Lewis, M. (1996) “Structure of the E. coli lactose operon repressor and its complexes with DNA and inducer,” Biophys. J. 70, TUAM5-TUAM5.
- Kercher, M.A., Chang, G., Horton, N.C., Lu, P., Miller, J.H., Pace, H.C. & Lewis, M. (1996) “Structure and genetics of the lactose operon repressor,” FASEB J. 10, 6-6.
- Lewis, M., Chang, G., Horton, N.C., Kercher, M.A., Pace, H.C., Schumacher, M.A., Brennan, R.G., & Lu, P. (1996) “Crystal structure of the lactose operon repressor and its complexes with DNA and inducer”, Science 271, 1201-1332.
- *Featured on the cover.
- Horton, N. & Lewis, M. (1992) “Calculation of the free energy of association for protein complexes”, Protein Science 1, 169-181.
- Structural Coordinates






