Associate Professor
Degrees and Appointments
- B.A. 2010, St. Olaf College
- Ph.D. 2013, University of Illinois at Urbana-Champaign
- Postdoctoral Researcher 2013-2016, University of Oxford
- Assistant Professor 2016, University of Arizona
- Associate Professor, University of Arizona, 2021
Field of Study: Biochemistry, Analytical Chemistry
Awards and Honors
- 2023 Distinguished Scholar Award, University of Arizona
- 2022 American Chemical Society Division of Analytical Chemistry Arthur F. Findeis Award for Achievements by a Young Analytical Scientist
- 2021 American Society for Mass Spectrometry Ron Hites Award
- 2021 Galileo Circle Curie Award, University of Arizona
- 2019 NSF CAREER Award
- 2019 Journal of the American Society for Mass Spectrometry Outstanding Reviewer Award
- 2018 American Society for Mass Spectrometry Research Award
- 2017 Bisgrove Scholar Award
Research Specialties: Bioanalytical, Biophysics, Chemical Biology, Instrument Development, Metabolism, Signaling, and Regulation, Protein and Membrane Biochemistry, Structural Biology
Research
Membrane proteins play a number of critical biochemical roles and make up the majority of drug targets. Despite their importance, membrane proteins remain challenging systems for analysis due to their amphipathic nature and low expression levels. Moreover, the lipid bilayer can play an important but largely unexplored role in regulating membrane protein structure and function. New analytical and biochemical methods are necessary to better understand and design drugs to target membrane proteins.
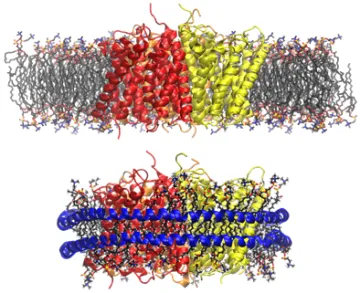
Membrane protein AmtB modeled in a POPC membrane (top) and a Nanodisc (bottom)
Research in the Marty lab is focused on developing mass spectrometry methods to study the structure and biophysics of membrane proteins and antimicrobial peptides. Working at the interface of Analytical Chemistry and Biochemistry, we use lipoprotein Nanodiscs to solubilize membrane proteins in a lipid bilayer with a defined composition. Nanodiscs are nanoscale discoidal lipid bilayers encircled by two amphipathic membrane scaffold proteins.
By using nondenaturing nano-electrospray ionization, we can keep the Nanodisc complex intact inside the mass spectrometer and gradually release the membrane protein with collisional activation. This approach, known as noncovalent or native mass spectrometry, allows us to measure the stoichiometry and lipid composition in large protein-lipid complexes to better understand protein-lipid interactions in membrane protein receptors and transporters. We are also using these approaches to study interactions of antimicrobial peptides with and within lipid bilayers.
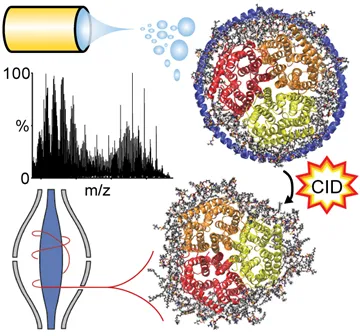
Following electrospray ionization, Nanodiscs are activated by gas-phase collision induced dissociation (CID) to release the membrane protein bound to dozens of lipids for high-resolution Orbitrap mass analysis, which results in a highly complex spectral pattern.
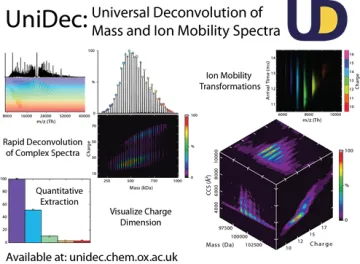
Because mass spectra of Nanodiscs are highly complex, we are interested in development of new computational approaches and software for analysis of native MS data. This work builds on UniDec, software we have developed to rapidly and robustly deconvolve mass and ion mobility spectra. We are also interested in developing cross-linking and covalent labeling approaches to use bottom-up proteomics to study the structure of membrane proteins in their native membrane environment.
We are also developing novel lipidomics experiments to measure affinity of lipids to membrane proteins. Here, we use LC-MS/MS to identify and quantify the lipids inside nanodiscs with and without membrane proteins, exploring enrichment of specific lipids in nanodiscs with a given membrane protein target. These experiments take advantage of cutting-edge lipidomics techniques, including ozone-induced dissociation to localize double bond positions.






