Associate Professor
Degrees and Honors
- B.S. 1983-1987, Bowdoin College
- Ph.D. 1987-1997, Yale University
- Postdoctoral Fellow 1994-2000, MIT
Awards and Honors
- Merck/MIT Fellow, 1998-1999
- Helen Hay Whitney Fellow, 1995-1998
- NSF Graduate Research Fellow, 1988-1991
Research
Molecular evolution is a critical research focus in the post-genome age of biology. With the large number of genome sequencing projects and the expansion of biomolecular structure databases, we have the opportunity in the 21st century to comprehensively appreciate and study the diversity and diversification of biological molecules. My research is focused on four areas within this field: 1) the origin and evolution of the approximately 1000 known basic types of protein structure, called folds; 2) the evolution of the function of sequence-specific DNA-binding proteins; 3) the evolution of proteins to avoid incorrect folding and aggregation, which can lead to many diseases, including Alzheimer's and prion disorders; 4) the evolution of protein toxins from nontoxic ancestor proteins. We approach these problems using a combination of computational studies (bioinformatics) with experimental structural biology (NMR and X-ray crystallography), biochemistry, and protein design.
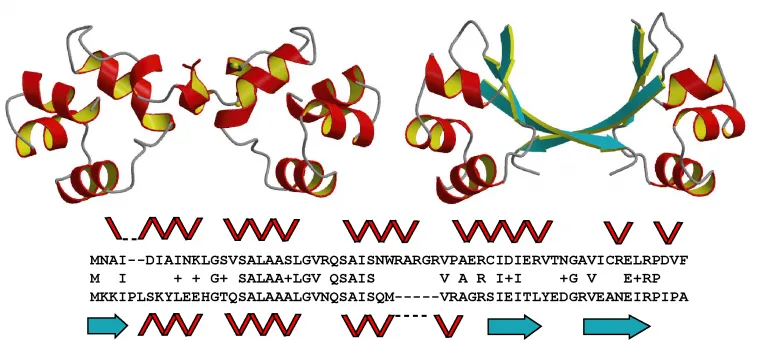
"Foldability of a natural de novo evolved protein," D. Bungard, J.S. Copple, J. Yan, J.J. Chhun, V.K. Kumirov. S.G. Foy, J. Masel, V.H. Wysocki, M.H.J. Cordes Structure (2017), in press.
"Spider, bacterial and fungal phospholipase D toxins make cyclic phosphate products," D.M. Lajoie, M.H.J. Cordes Toxicon 108, 176-180 (2015). PMID: 26482933
"The recent de novo origin of protein C-termini," M.E. Andreatta, J.A. Levine, S.G. Foy, L.D. Guzman, L.J. Kosinski, M.H.J. Cordes, J. Masel Genome Biol Evol 21, 1686-701 (2015). PMID: 26002864
"Studying protein fold evolution with hybrids of differently folded homologs," K.V. Eaton, W.J. Anderson, M.S. Dubrava, V.K. Kumirov, E.M. Dykstra, M.H.J. Cordes Protein Eng Des Sel , 28, 241-250 (2015). PMID: 25991865
"Variable substrate preference among phospholipase D toxins from sicariid spiders," D.M. Lajoie, S.A. Roberts, P.A. Zobel-Thropp, J.L. Delahaye, V. Bandarian, G.J. Binford, M.H.J. Cordes J. Biol. Chem. 290, 10994-1007 (2015). PMID: 25752604
"Phospholipase D toxins of brown spider venom convert lysophosphatidylcholine and sphingomyelin to cyclic phosphates," D.M. Lajoie, P.A. Zobel-Thropp, V.K. Kumirov, V. Bandarian, G.J. Binford, M.H.J. Cordes PLoS ONE 8, e72372 (2013). PMID: 24009677
"A role for indels in the evolution of Cro protein folds," K.L Stewart, M.R. Nelson, K.V. Eaton, W.J. Anderson, M.H.J. Cordes, Proteins 81, 1988-1996 (2013). PMID: 23843258
"A polymetamorphic protein," K.L. Stewart, E.D. Dodds, V.H. Wysocki, M.H.J Cordes, Protein Sci., 22, 641-649 (2013). PMID: 23471712
"Reengineering Cro protein functional specificity with an evolutionary code", B.M. Hall, E.E Vaughn, A.R. Begaye, M.H.J. Cordes, J. Mol. Biol. 413, 914-928 (2011). PMID: 21945527
"Evolutionary bridges to new protein folds: design of C-terminal Cro protein chameleon sequences", W.J. Anderson, L.O. Van Dorn, W.M. Ingram, M.H.J. Cordes, Protein Eng. Des. Sel. 24, 765-775 (2011). PMID: 21676898.
"Determinants of gas-phase disassembly in homodimeric protein complexes with related yet divergent structures", E.D. Dodds, A.E. Blackwell, C.M. Jones, K.L. Holso, D.J. O'Brien, M.H.J. Cordes, V.H. Wysocki, Anal. Chem. 83, 3881-3889 (2011). PMID: 21486017
"Measuring (1H) (N) temperature coefficients in invisible protein states by relaxation dispersion NMR spectroscopy", G. Bouvignies, P. Vallurupalli, M.H.J. Cordes, D.F. Hansen, L.E. Kay, J. Biomol. NMR, 50, 13-18 (2011). PMID: 21424227
"A simple method for measuring signs of (1)H (N) chemical shift differences between ground and excited protein states", G. Bouvignies, D.M. Korzhnev, P. Neudecker, D.F. Hansen, M.H.J. Cordes, L.E. Kay, J.Biomol. NMR, 47, 135-141 (2010). PMID: 20428928
"Molecular evolution and proposed nomenclature of the gene family that includes sphingomyelinase D in Sicariid spider venoms", G.J. Binford, M.R. Bodner, M.H.J. Cordes, K.L. Baldwin, M.R. Rynerson, S.N. Burns, and P.A. Zobel-Thropp, Mol. Biol. Evol., 26, 547-566 (2009). PMID: 19042943
"Structural features and the persistence of acquired proteins", H.P. Narra, M.H.J. Cordes, and H. Ochman, Proteomics, 8, 4772-4781 (2008). PMID: 18924109
"N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces", M.S. Dubrava, W.M. Ingram, S.A. Roberts, A. Weichsel, W.R. Montfort, and M.H.J. Cordes, Protein Science, 17, 793-802(2008). PMID 18369196
"Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds", C.G. Roessler, B.M. Hall, W.J. Anderson, W.M. Ingram, S.A. Roberts, W.R. Montfort, and M.H.J. Cordes, Proc. Natl. Acad. Sci. USA, 2343-8 (2008). See also commentary, p. 2759. PMID: 18227506
"Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA", B.M. Hall, S.A. Roberts, A.M. Heroux, and M.H.J. Cordes, J. Mol. Biol, 375, 802-811 (2008). PMID: 18054042
"Conservation and diversity in the immunity regions of wild phage with the immunity specificity of phage lambda", P.H. Degnan, C.B. Michalowski, A.C. Babic, M.H.J. Cordes, and J.W Little, Mol. Microbiol, 64, 232-244 (2007). PMID: 17376085
"High polar content of long buried blocks of sequence in protein domains suggests selection against amyloidogenic nonpolar sequences", A.U. Patki, A.C. Hausrath and M.H.J. Cordes, J. Mol. Biol., 362, 800-809 (2006). PMID: 16935301
"Relationship between sequence determinants of stability for two natural homologous proteins with different folds", L.O. Van Dorn, T. Newlove, S. Chang, W.M. Ingram and M.H.J. Cordes, Biochemistry, 45, 10542-10553 (2006). PMID: 16939206
"A trade between similar but nonequivalent intrasubunit and intersubunit contacts in Crodimer evolution", T. Newlove, K.R. Atkinson, L.O. Van Dorn, and M.H.J. Cordes, Biochemistry, 45, 6379-6391 (2006). PMID: 16700549
"Lateral gene transfer of a dermonecrotic toxin between spiders and bacteria", M.H.J. Cordes and G.J. Binford, Bioinformatics, 22, 264-268 (2006). [first and co-corresponding author] PMID: 16332712
"Sequence determinants of a conformational switch in a protein structure", T.A. Anderson, M.H.J. Cordes, and R.T. Sauer, Proc. Natl. Acad. Sci. U.S.A., 102, 18344-18349 (2005). PMID: 16344489
"Sequence correlations between Cro recognition helices and cognate O R consensus half-sites suggest conserved rules of protein-DNA recognition", B.M. Hall, K.R. LeFevre, and M.H.J. Cordes, J. Mol. Biol., 350, 667-681 (2005). PMID: 15967464
"Sphingomyelinase D from venoms of brown spiders: evolutionary insights from cDNA sequences and gene structure", G.J. Binford, M.H.J. Cordes, and M.A. Wells, Toxicon, 45, 547-560 (2005). PMID: 15777950
"Secondary structure switching in Cro protein evolution", T. Newlove, J. H. Konieczka, and M.H.J. Cordes, Structure, 12, 569-581 (2004) (editor's featured article). PMID: 15062080
"Retroevolution of lambda Cro toward a stable monomer", K.R. LeFevre,and M.H.J. Cordes, Proc. Natl. Acad. Sci. U.S.A., 100, 2345-2350 (2003). PMID: 12598646
"Solution structure of switch Arc, a mutant with 310 helices replacing a wild-type ß-ribbon," M.H.J. Cordes, N.P. Walsh, C.J. McKnight, R.T. Sauer, Journal of Molecular Biology, 326, 899-909 (2003). PMID: 12581649
"An Evolutionary Bridge to a New Protein Fold," M.H.J. Cordes, R.E. Burton, N.P. Walsh, C.J. McKnight, R.T. Sauer, Nature Structural Biology, 7, 1129-1132 (2000). PMID: 11101895
"Evolution of a Protein Fold In Vitro," M.H.J. Cordes, N.P. Walsh, C.J. McKnight, R.T. Sauer, Science, 284, 325-327(1999). PMID: 10195898
"Tolerance of a Protein to Multiple Polar-to-hydrophobic Surface Substitutions," M.H.J. Cordes, R.T. Sauer, Protein Sci, 8, 318-325 (1999). PMID: 10084325
for measuring signs of (1)H (N) chemical shift differences between ground and excited protein states", G. Bouvignies, D.M. Korzhnev, P. Neudecker, D.F. Hansen, M.H.J. Cordes, and L.E. Kay, J.Biomol, NMR, 47, 135-141 (2010). PMID: 20428928
"Molecular evolution and proposed nomenclature of the gene family that includes sphingomyelinase D in Sicariid spider venoms", G.J. Binford, M.R. Bodner, M.H.J. Cordes, K.L. Baldwin, M.R. Rynerson, S.N. Burns, and P.A. Zobel-Thropp, Mol. Biol. Evol., 26, 547-566 (2009). PMID: 19042943
"Structural features and the persistence of acquired proteins", H.P. Narra, M.H.J. Cordes, and H. Ochman, Proteomics, 8, 4772-4781 (2008). PMID: 18924109
"N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces", M.S. Dubrava, W.M. Ingram, S.A. Roberts, A. Weichsel, W.R. Montfort, and M.H.J. Cordes, Protein Science, 17, 793-802(2008). PMID 18369196
"Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds", C.G. Roessler, B.M. Hall, W.J. Anderson, W.M. Ingram, S.A. Roberts, W.R. Montfort, and M.H.J. Cordes, Proc. Natl. Acad. Sci. USA, 2343-8 (2008). See also commentary, p. 2759. PMID: 18227506
"Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA", B.M. Hall, S.A. Roberts, A.M. Heroux, and M.H.J. Cordes, J. Mol. Biol, 375, 802-811 (2008). PMID: 18054042
"Conservation and diversity in the immunity regions of wild phage with the immunity specificity of phage lambda", P.H. Degnan, C.B. Michalowski, A.C. Babic, M.H.J. Cordes, and J.W Little, Mol. Microbiol, 64, 232-244 (2007). PMID: 17376085
"High polar content of long buried blocks of sequence in protein domains suggests selection against amyloidogenic nonpolar sequences", A.U. Patki, A.C. Hausrath and M.H.J. Cordes, J. Mol. Biol., 362, 800-809 (2006). PMID: 16935301
"Relationship between sequence determinants of stability for two natural homologous proteins with different folds", L.O. Van Dorn, T. Newlove, S. Chang, W.M. Ingram and M.H.J. Cordes, Biochemistry, 45, 10542-10553 (2006). PMID: 16939206
"A trade between similar but nonequivalent intrasubunit and intersubunit contacts in Crodimer evolution", T. Newlove, K.R. Atkinson, L.O. Van Dorn, and M.H.J. Cordes, Biochemistry, 45, 6379-6391 (2006). PMID: 16700549
"Lateral gene transfer of a dermonecrotic toxin between spiders and bacteria", M.H.J. Cordes and G.J. Binford, Bioinformatics, 22, 264-268 (2006). PMID: 16332712
"Sequence determinants of a conformational switch in a protein structure", T.A. Anderson, M.H.J. Cordes, and R.T. Sauer, Proc. Natl. Acad. Sci. U.S.A., 102, 18344-18349 (2005). PMID: 16344489
"Sequence correlations between Cro recognition helices and cognate O R consensus half-sites suggest conserved rules of protein-DNA recognition", B.M. Hall, K.R. LeFevre, and M.H.J. Cordes, J. Mol. Biol., 350, 667-681 (2005). PMID: 15967464
"Sphingomyelinase D from venoms of brown spiders: evolutionary insights from cDNA sequences and gene structure", G.J. Binford, M.H.J. Cordes, and M.A. Wells, Toxicon, 45, 547-560 (2005). PMID: 15777950
"Secondary structure switching in Cro protein evolution", T. Newlove, J. H. Konieczka, and M.H.J. Cordes, Structure, 12, 569-581 (2004). PMID: 15062080 (Editor's featured article, must read on Faculty of 1000).
"Retroevolution of lambda Cro toward a stable monomer", K.R. LeFevre,and M.H.J. Cordes, Proc. Natl. Acad. Sci. U.S.A., 100, 2345-2350 (2003). PMID: 12598646






